Educational game reviewing the processes of the Citric Acid Cycle (aka. TCA, CAC, or Krebs Cycle) and helps visualize the relevant molecules and their bonds in 3D space.
Created in Unity3D, coded in C#.
Created with Tom Moreton as part of a Master's course in Interactive Visualisation at the Glasgow School of Art.
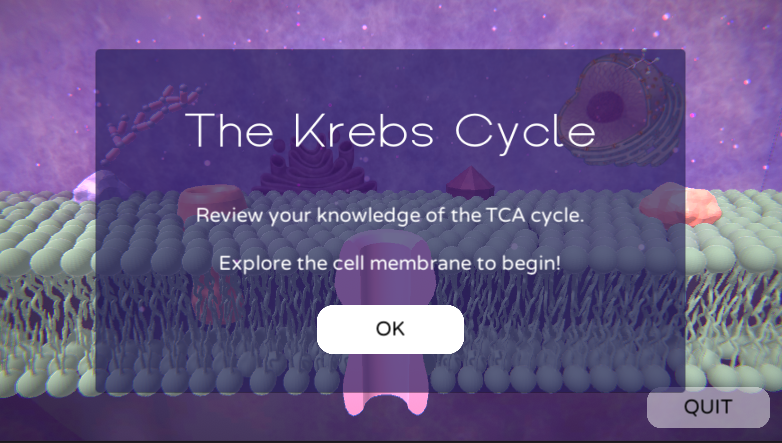

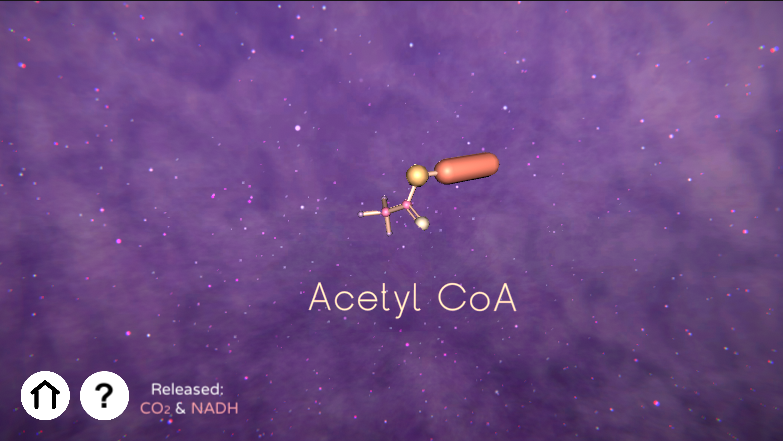
GameMode

Moodboard

Building Intro Scene

Mapping UI panels
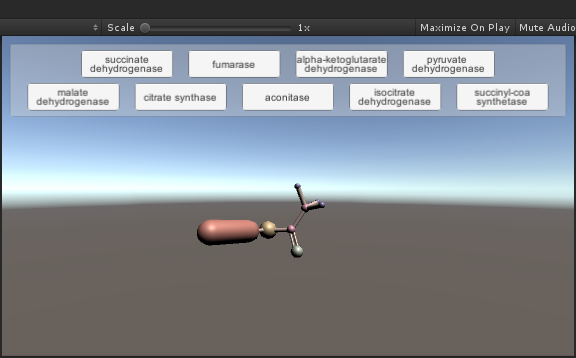
Importing 3D Models
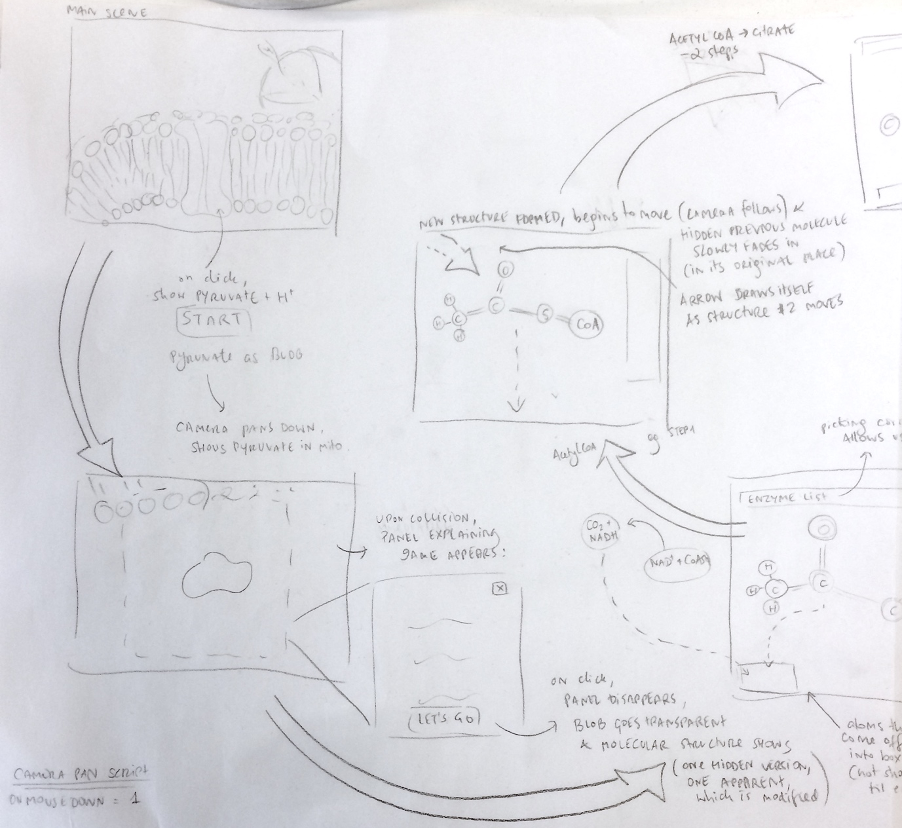
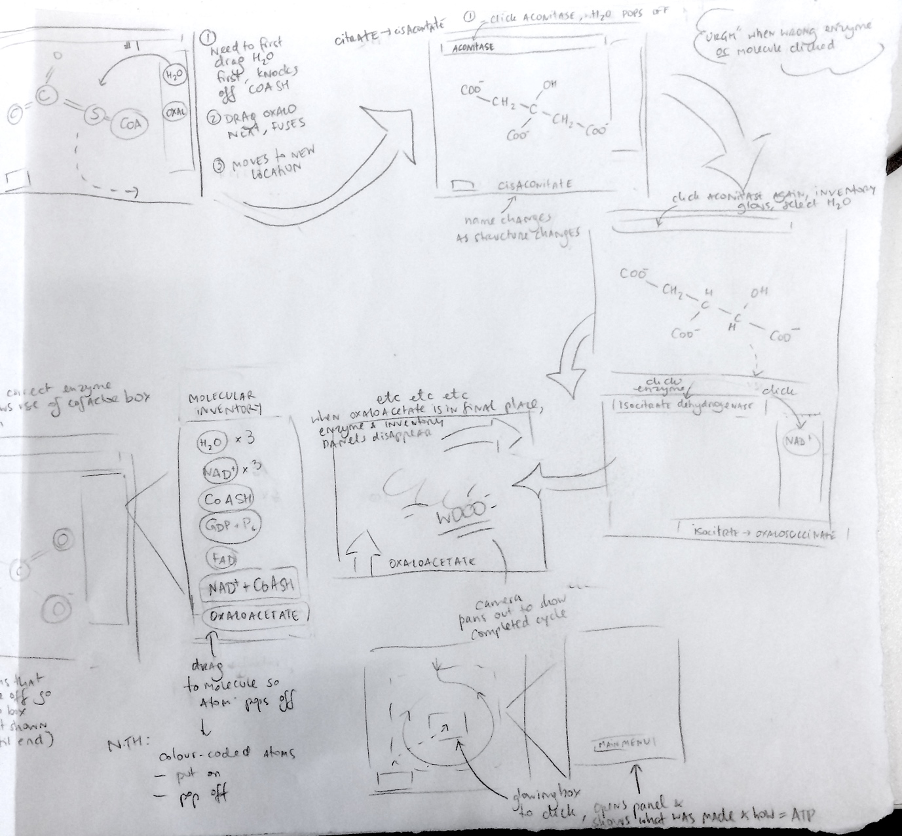
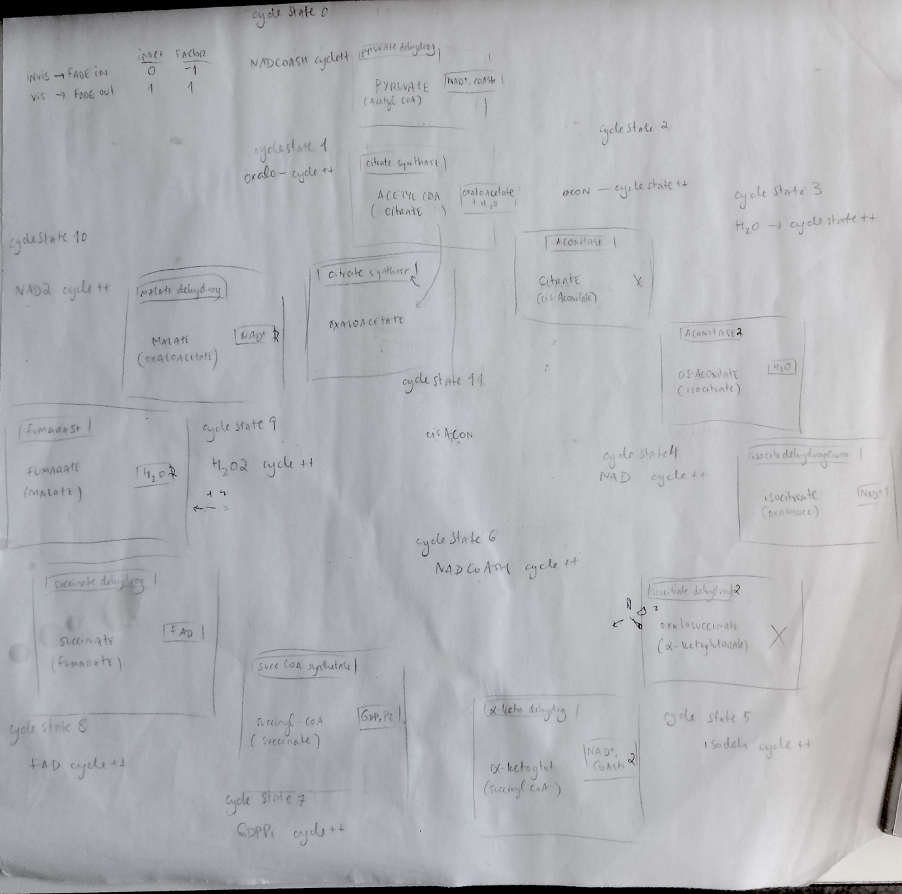
Mapping the cycle
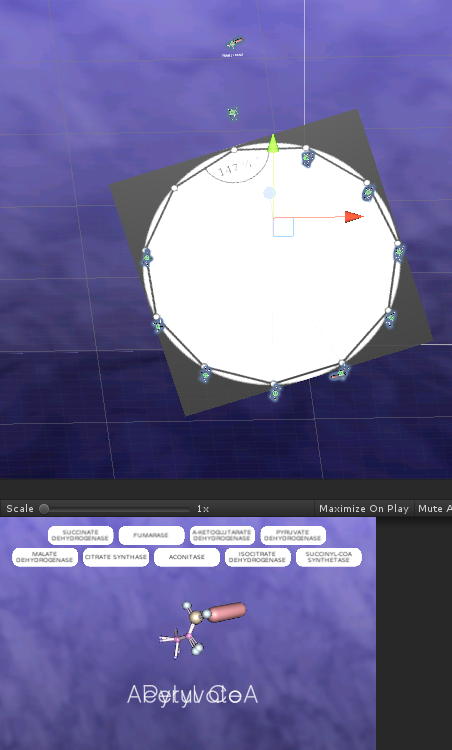
Mapping the cycle
View more 3D Work
Anatomy of FGM/C Webapp / Anatomical 3D Models / Volumetric Visualisation / Krebs Cycle Game

